MetaboLabPy
The Graphical User Interface
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
The main window of the MetaboLabPy GUI is organised as several tabs, each of which focusing on a specific area associated with the loaded NMR spectra. The first tab (NMR Spectrum) displays the selected NMR spectra. The Processing Parameters tab is used to set data processing such as phase correction values, the amount of line broadening etc. The information set in the Display Parameters tab determines how the NMR spectra are displayed (e.g. plot colour, minimum and maximum contour levels for 2D NMR spectra). The Acquisition Parameters tab summarises settings used to acquire the NMR spectrum. Information saved in title files is presented in the Title file tab. The used pulse program is shown inside the pUlseProgram tab. Reading NMR data from raw spectrometer data files as well as NMR data processing can be automated using scripts displayed in the Script tab. Any console output including error messages will be displayed in the cOnsole tab. Help files as well as links to popular NMR data bases such as BML, HDMB, KEGG and more are available in the Help tab. Every tab can be activated using keyboard shortcuts. On the Windows platform this is the combination of Ctrl + ⇧ + the first capitalised letter of the tab name (e.g. Ctrl + ⇧ + D to reach the Display Parameters tab and Ctrl + ⇧ + O to reach the cOnsole tab). Specifics for every tab are discussed below. There are specific action Items available from the different menus. These are discussed in the Menu Items section below.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
Once NMR spectra are loaded into the software, additional options appear below the spectrum display. For 1D-NMR spectra these options include Data Pre-Processing, Peak Picking and Spline baseline correction. These options will be discussed in Basic NMR Data Processing.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
In this tab, NMR processing parameters can be seen for each spectrum. These include the amount of zero filling, the type of window function, amount of line broadening, phase correction values to ensure pure absorptive lineshapes and whether to perform post-acquisition water suppression and strip-transform if necessary. The baseline correction parameters can be determined automatically, which is covered in Basic NMR Data Processing. Similarly, phase correction parameters are best to be determined interactively, which is also covered in Basic NMR Data Processing.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
Settings in this tab determine the look and feel of the displayed NMR spectra. Adjustable parameters include line colour, which for 1D-NMR spectra is set as PosCol and for 2D-NMR spectra is both PosCol and NegCol, PosCol for positive contour lines and NegCol for negative contour lines. The user can choose RGB and then provide RGB values in the interval 0 to 1, or colours which are labelled in the drop down menus.
2D-NMR spectra are displayed as contour plots, the number of contour levels (nLevels) as well as the minimum and maximum contour levels can be set here. The axis type can be set to Hz or ppm, axis labels selected and a phase reference dataset can be set, which is usually experiment 1 and the colour of that phase reference spectrum can be set when in interactive phase correction mode to ensure consistent phase corrections across a large set of 1D-NMR spectra.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
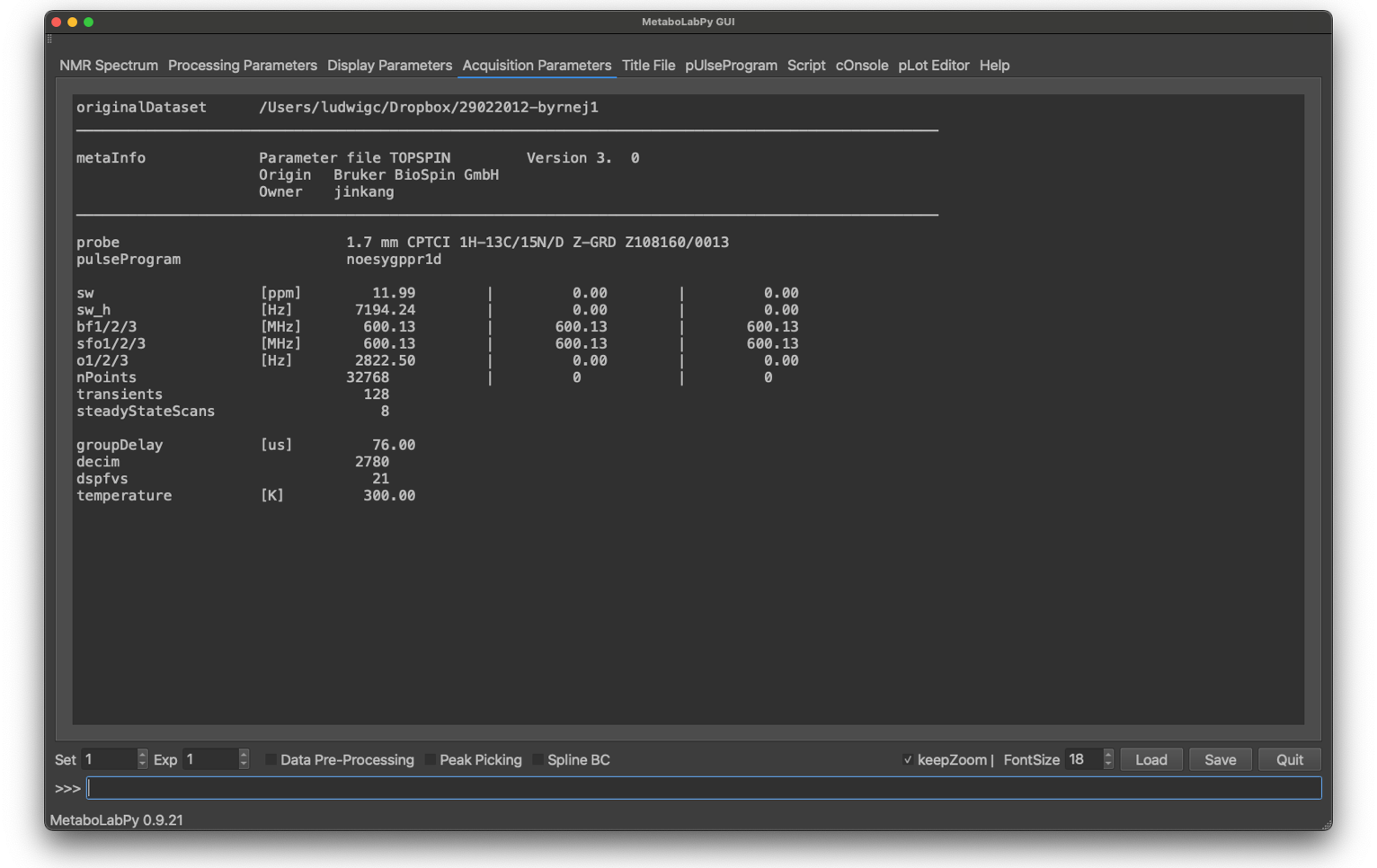
The Acquisition Parameters tab is display only. The most important acquisition parameters are displayed here and are here for reference purposes.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
Both Bruker and Varian allow for meta-information to be associated with each NMR spectrum at time of acquisition to uniquely identify every single NMR spectrum. It is the responsibility of the NMR spectrometer operator to ensure that this information is associated with each spectrum, although MetaboLabPy also contains functionality to associate that meta-information with each NMR spectrum from an Excel spreadsheet, although this requires the use of the automatic sample changing robot SampleJet and therefore is entirely Bruker centric.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
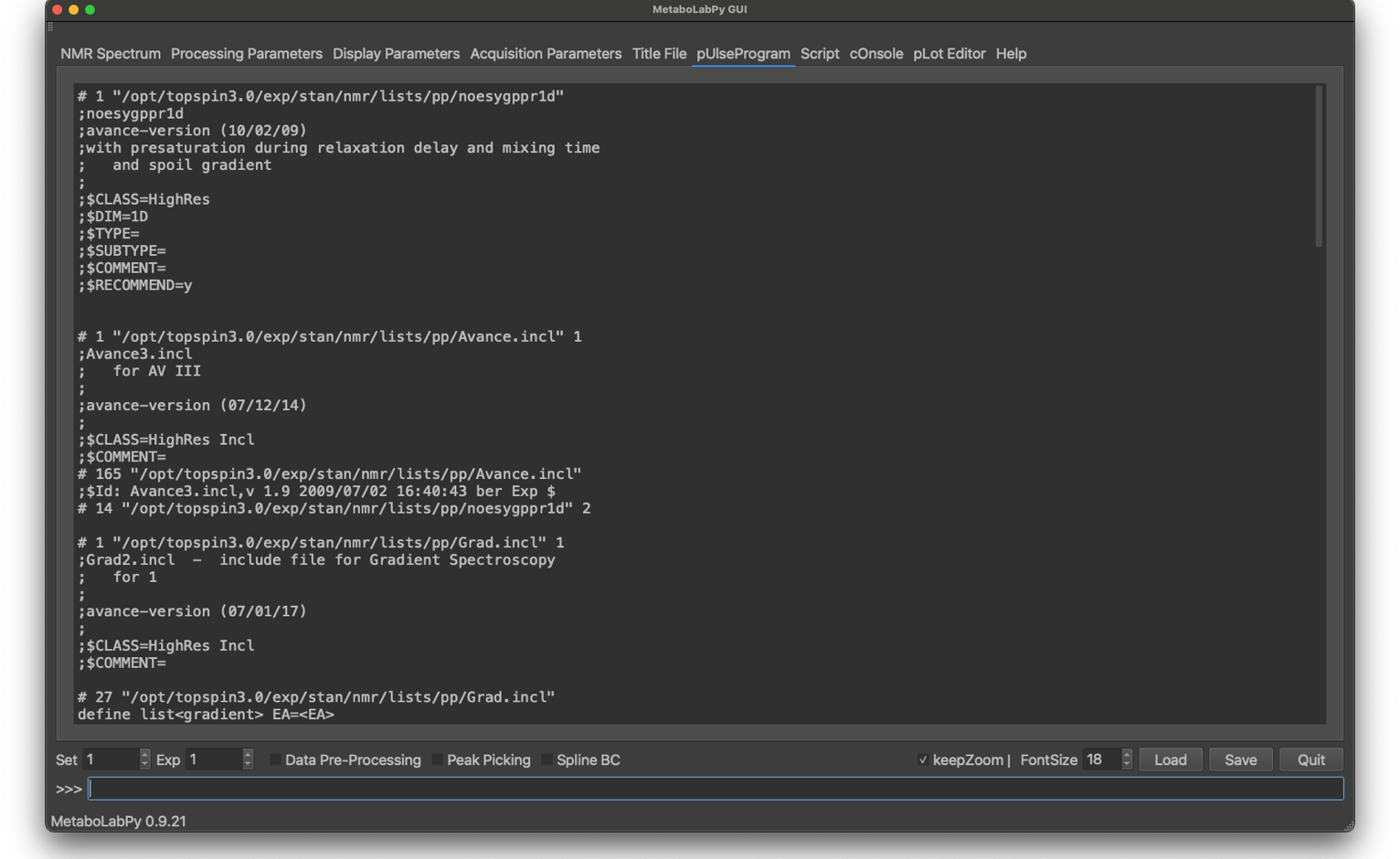
The TopSpin software stores a version of the pulse sequence used to acquire the NMR spectrum with each raw NMR data file. MetaboLabPy reads this pueseprogram in and displays the pueseprogram in this tab.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
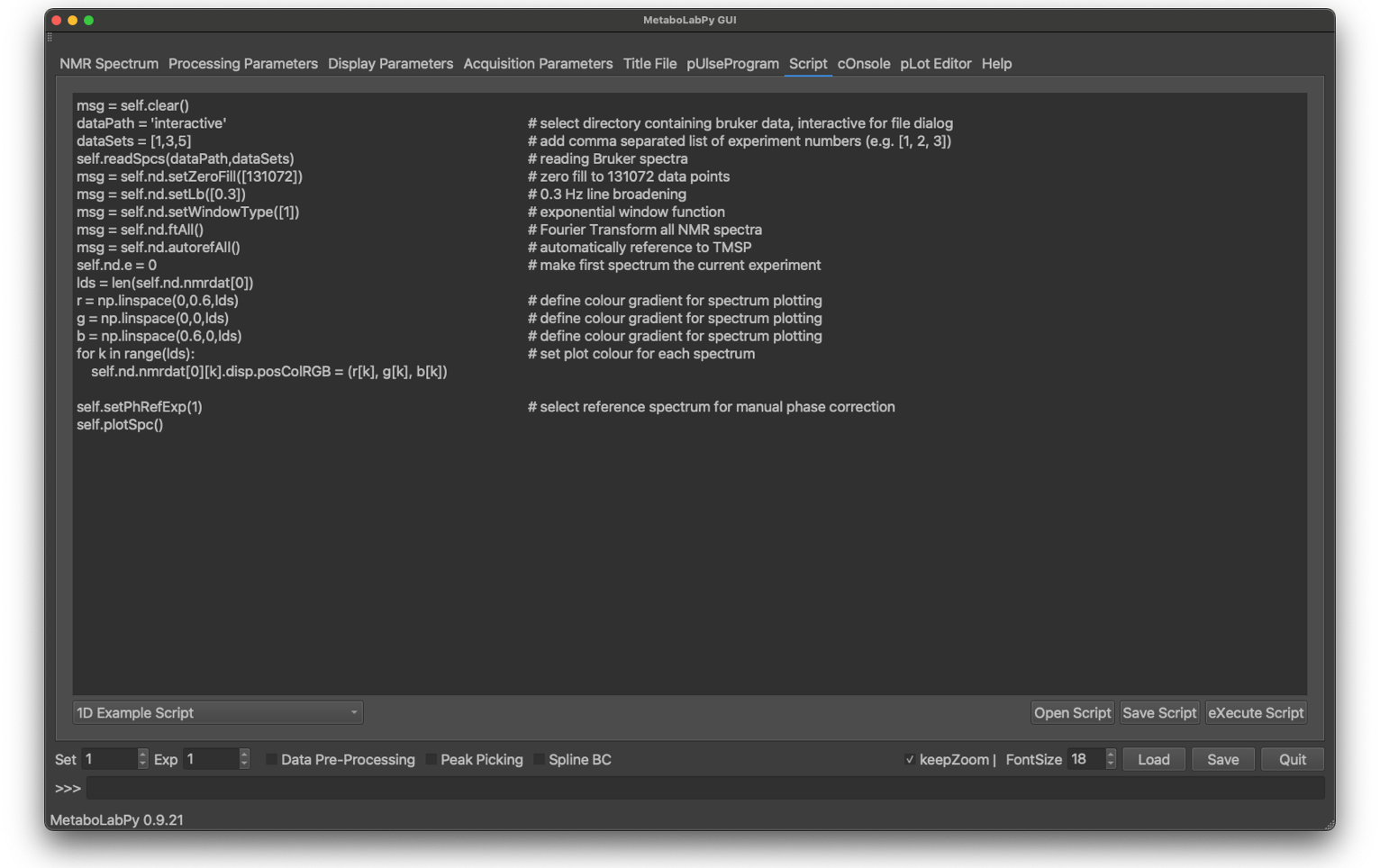
Because MetaboLabPy is implemented as a series of python classes, procedures can be easily automated via python scripts. MetaboLabPy has a scripting interface and a collection of example scripts, all of which are well documented so that use of them should be more or less self-explanatory. Especially reading in a series of NMR spectra is greatly facilitated using the scripting interface.
Scripting is also possible just running the python interpreter and creating a NmrDataSet object or by doing the same as part of a Jupyter notebook. Both of these possibilities will be discussed in MetaboLabPy Scripts and MetaboLabPy and Jupyter Notebooks, respectively.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
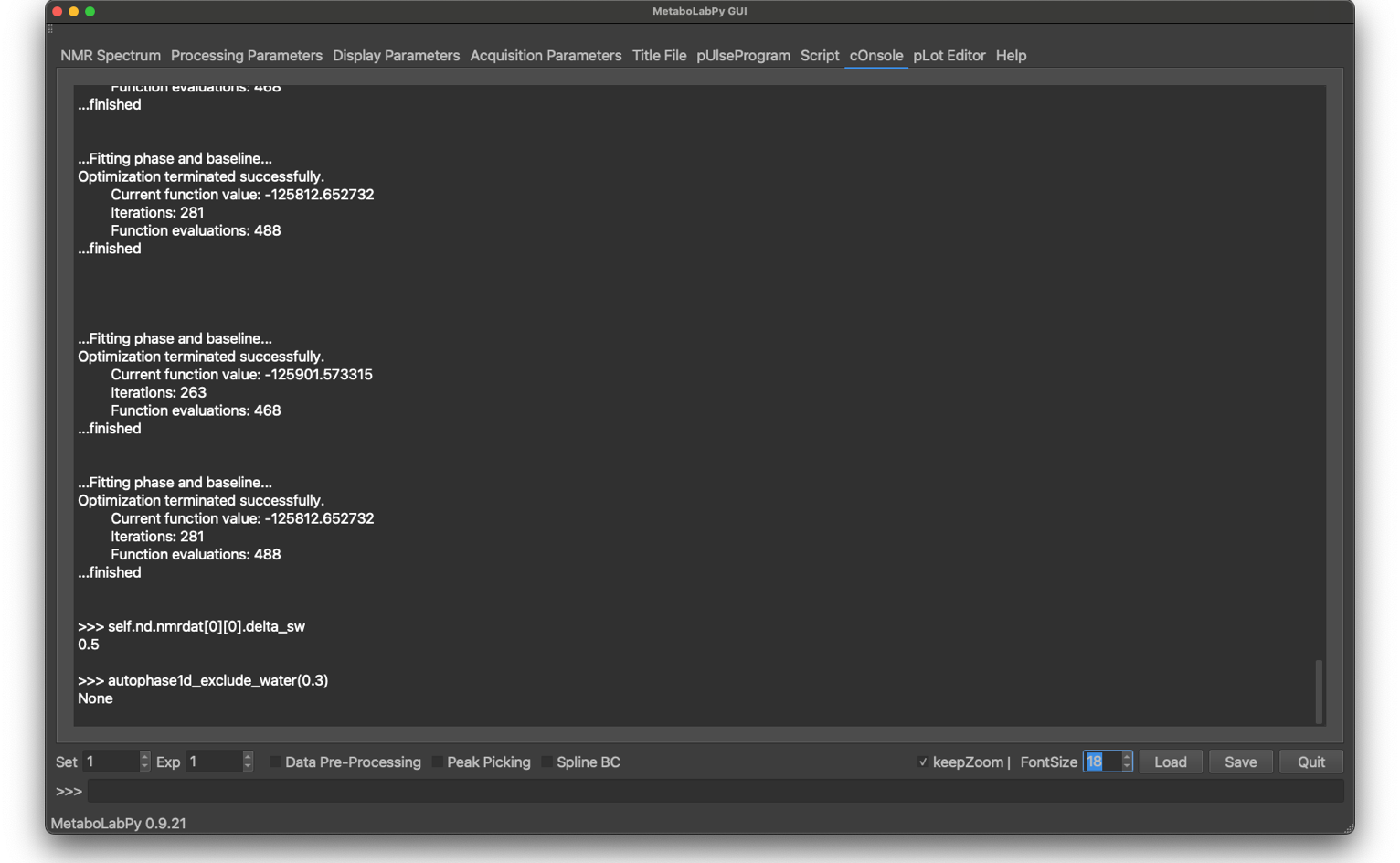
Most GUI routines re-route their standard output device to the cOnsole tab. Any output will be displayed in this window. Execute script texts will appear in blue, error messages in red and normal output either in black or in white depending on light or dark mode settings.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
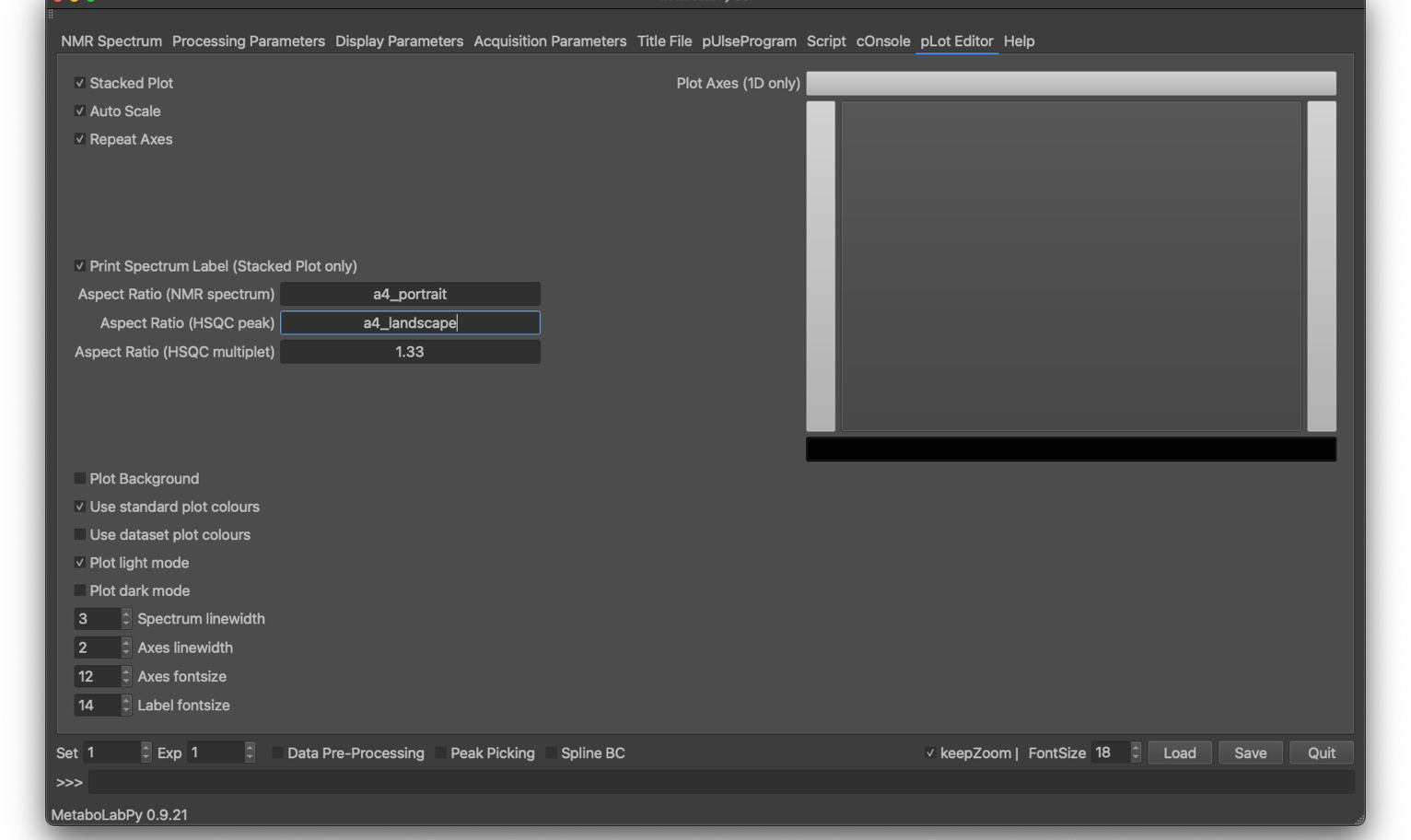
PDF plots of NMR spectra can be generated in MetaboLabPy by choosing the option “Print” in the “Command Line” menu, or by using the keyboard shortcut Ctrl + P (⌘ + P in macOS).
This tab controls the layout and look-and-feel of PDF plots of 1D- and 2D-NMR spectra. If multiple spectra are displayed, they can be plotted as overlaid spectra (with the stacked plot option turned off, or as stacked plot when this option is selected. When spectra are plotted as stacked, the axes selected can be repeated or kept only for the first spectrum (plotted on the bottom).
Spectra can also be scaled to maximum vertical size if the auto scale option is selected. The axes to be plotted (bottom, left, top, right) can be selected graphically. The background can be transparent or plotted (white in light mode, dark gray in dark mode). Spectrum plot colours can either be the same as on-screen or MetaboLabPy standard plot colours, which is, in particular, useful if the PDF plot mode (which can be set to either light or dark mode) is different from the GUI colour mode. Finally, the line widths for the spectrum and the axes can be controlled, as can font sizes for axes and labels.
In addition to font sizes and line widths, the aspect ratio can be controlled. If the ‘auto’ is select, the aspect ratio will be as displayed within the graphical user interface. Alternatively the aspect ratio can be specified with a floating point number. E.g. choose 1.33 to set the aspect ratio (width / height) to 4/3.
In order to generate a pdf file with A4 dimensions, the aspect ratios can be set to a4_portrait or a4_landscape as well. In case of a stack plot, this then refers to the dimensions of the finished pdf file.
Each spectrum in a stacked spectrum plot can be automatically annotated. To plot this spectrum label, the following line needs to be included in the title file of the NMR spectrum:
spcLabel: label
Where label needs to be replace with the spectrum label to be printed in the stacked plot.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
As the name suggests, this tab contains the included documentation, which is also available at https://ludwigc.github.io/metabolabpy/index.html. In addition quick access to NMR and metabolism related databases, including the Birmingham Metabolite Library (BML), the Human Metabolome Database (HMDB), the Small Molecule Pathway Database (SMPDB), the Biological Magnetic Resonance Bank Database (BMRB), the Kyoto Encyclopedia of Genes and Genomes Database (KEGG), NMRShiftDB2, the Database for Organic Compounds (SDBS) and the SpinCouple Database.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
To explore the different options, click on the different menu items. Click on the X to close the pop-up window.
NMR Spectrum | Processing Parameters | Display Parameters | Acquisition Parameters | Title File | pUlseProgram | Script | cOnsole | pLot Editor | Help | Menu Items
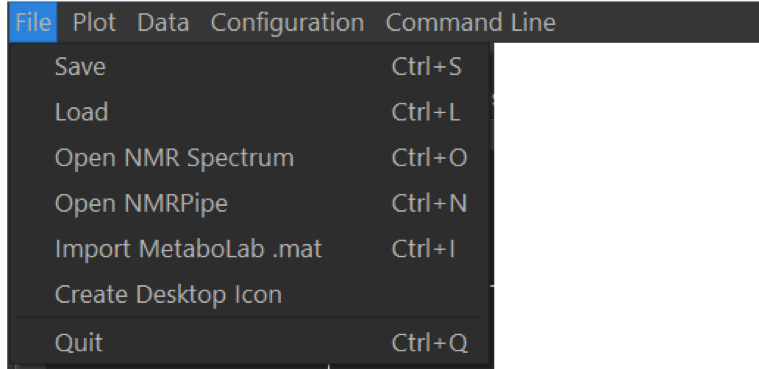
NMR data can be read into MetaboLabPy from different sources. Both Bruker and Varian raw NMR data can be read into MetaboLabPy using the Open NMR Spectrum entry. On Windows, this can be achieved using the keyboard shortcut Ctrl + O, on macOS using ⌘ + O. NMR data can be save using the MetaboLabPy data file format using Ctrl + S (⌘ + S on macOS). Files saved in this format can be loaded in again using Load (Ctrl + L on Windows, ⌘ + S on macOS).
Data saved in the Matlab based MetaboLab software as a .mat file, can be imported into MetaboLabPy using the keyboard shortcut Ctrl + I (⌘ + I on macOS). Any data preprocessing options are imported as well.
On Windows a desktop icon capable of starting MetaboLabPy can be generated with the Create Desktop Icon entry. On macOS the equivalent LaunchPad starter can be generated by clicking on Create Launchpad Icon instead. The software can be shutdown by clicking on Quit (Ctrl + Q or ⌘ + Q).
In this menu, default configurations can be stored (Save as Default), reload and reset to MetaboLabPy defaults. The colour scheme can be set to system mode (same as operating system or enforce either high or dark mode. The application can be put into fullscreen/windowed mode (Alt + F). The splash screen can also be displayed for 5 seconds.
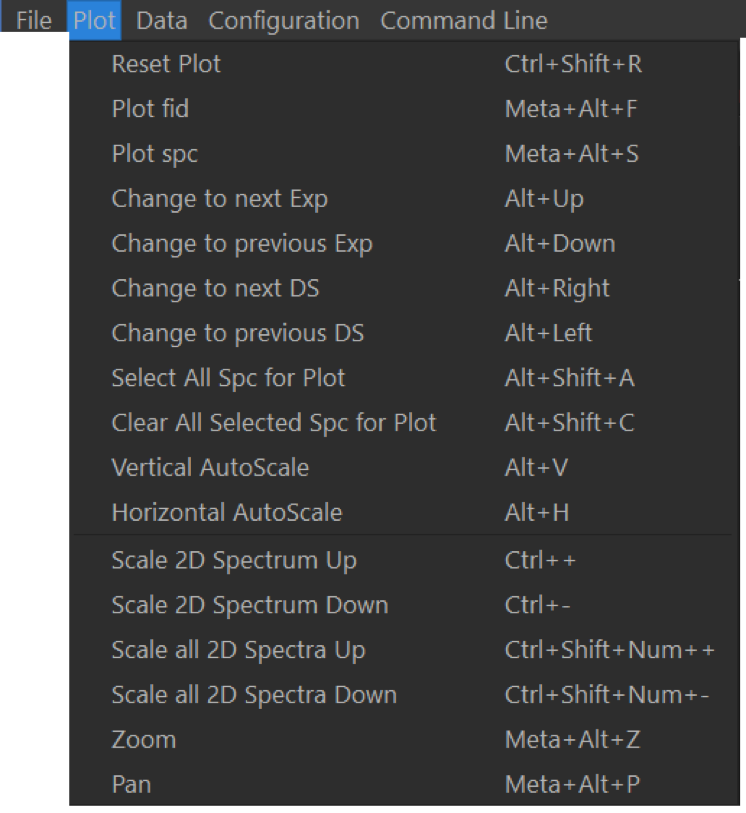
Actions in this menu are all related to NMR spectra display. Plot settings can be reset with Reset Plot (Ctrl + R or ⌘ + R). Spectrum display can be changed to the previous/next experiment with Alt +
To plot all spectra in the current dataset, click on Select All Sec for Plot (Alt + ⇧ + A), revert back to display the current spectrum only by clicking on Clear All Selected Spa for Plot (Alt + ⇧ + C).
For any spectral area, clicking on Vertical AutoScale (Alt + V) will change the vertical scaling to be optimal for the currently selected horizontal section of the NMR spectra. Clicking on Horizontal AutoScale (Alt + H) will unzoom the chemical shift axis but keep the current vertical scaling.
Mouse actions can be changed between zoom (Meta + Alt + Z, use right mouse button to unzoom) and pan (Meta + Alt + P), where the spectrum can be dragged across the screen.
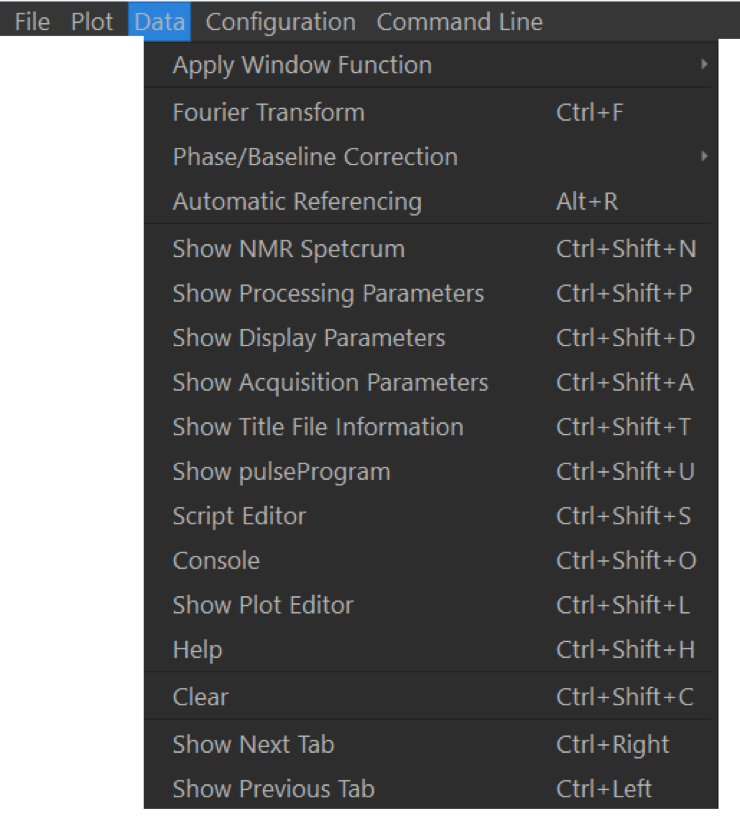
This menu deals with data processing options. Data can be Fourier-Transformed (Ctrl + F / ⌘ + F, automatically referenced to either TMSP or, if not present, to the water resonance in the middle of the spectrum (Alt + R / ⌘ + R). This menu also shows the keyboard shortcuts to access the different tabs of the GUI. All NMR data can be removed from memory via the Clear command (Ctrl + ⇧ + C / ⌘ + ⇧ + C).
The integrated command line can be (de-)activated by pressing escape. The command line keeps a history of executed commands, which can be displayed using the command h() on the command line. Previous commands can be recalled using